- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
GENE STRANDEDNESS OVERVIEW: 5 PRIME TO 3 PRIME
In this basic overview, we will be looking at how to distinguish between the 5' end and 3' end for any given gene on the browser. We will also look at identifying other general key components of genes on the Browser.
The 5' and 3' ends of a DNA or RNA molecule refer to the number of the carbon atom in a deoxyribose sugar molecule to which a phosphate group bonds. Transcription starts from the 5' end of the gene, making mRNA by extending the 3' end. Genes can be transcribed from the genomic DNA in either direction within the browser, left to right or right to left. Thus, the direction of transcription (and therefore translation) is arbitrary from the genomic point of view, with no apparent preference for either direction. Knowing which end of a given strand you are looking at is useful when trying to locate certain nucleotide sequences (e.g., start of transcription or start and stop of translation) or exons on a gene. To see a detailed example diagram and read a more in-depth explanation of directionality, click on the following link: Directionality (Molecular Biology).
In Figure 1, we have our example gene, MYC. If you look closely at the gene, you will see the blocks for exons and pointed dashed arrows for introns. Introns are removed from mRNAs after transcription, so the mature mRNA aligns to the genomic DNA in exon blocks. Hovering your mouse over any of the exons in a given gene will allow you to see the exon's number ranking in order within the gene — a useful tool for locating the 5' and 3' ends of a gene. Exon 1 is at the 5' end of the gene. In the intron spaces, the arrows point in the direction of transcription. Thus, for this gene, MYC's 5' end is on the left, and its 3' end is on the right.
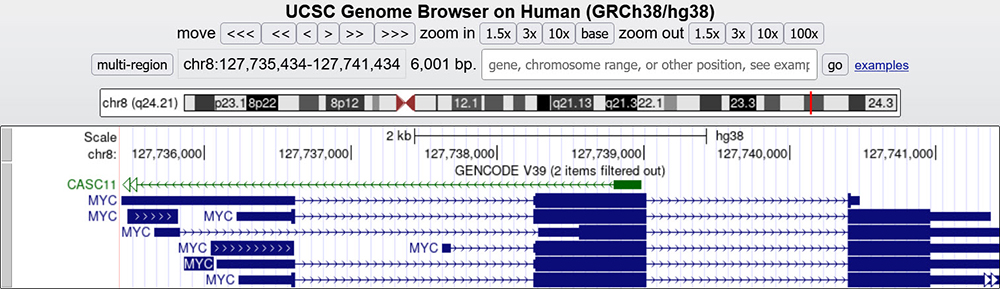
Figure 1. MYC gene main assembly window with exons and introns. The direction of the small intron arrows can be seen to go from left to right in the MYC gene, denoting the direction of the gene and signifying that the 5' end is to the left and 3' to the right ("top" strand).
Another similar method for figuring out the direction of a strand is by zooming out entirely on a gene and looking for the small blue rectangular parts at either end of the gene. In Figure 2, by completely zooming out and positioning the MYC gene in the frame, we can observe the two skinnier boxes, known as the non-coding or untranslated regions (UTR)
(
 http://genome.ucsc.edu/s/education/hg38_MYC).
By hovering our cursor over the skinnier boxes, a small text box will tell us the exon number preceding or following the skinny box (red circles).
One of them (at the 5' end) will be identified as Exon 1.
http://genome.ucsc.edu/s/education/hg38_MYC).
By hovering our cursor over the skinnier boxes, a small text box will tell us the exon number preceding or following the skinny box (red circles).
One of them (at the 5' end) will be identified as Exon 1.
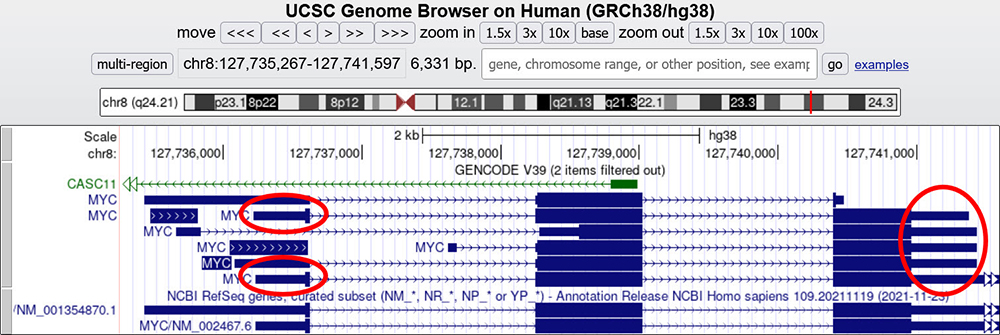
Figure 2. MYC gene main assembly window with exons and introns. In this picture, the skinny UTR region boxes are circled in red. Hovering over these regions will show the exon number for the gene, helping determine the overall direction when arrowheads are not present or when offscreen.
Our last example will look at a gene on the bottom strand/opposite strand. In Figure 3, the METTL3 gene has arrows facing in the opposite direction (right to left). Hovering over the exons, the furthermost exon to the right is
numbered "1 of 11" while the furthermost exon to the left is "11 of 11"
(
 http://genome.ucsc.edu/s/education/hg38_revStrandGene).
Zooming in to the amino acids within the exons of the METTL3 gene, we can see the numbering is backwards, going from right to left. This can be seen in Figure 4 and is also another useful technique in determining gene and strand direction
(
http://genome.ucsc.edu/s/education/hg38_revStrandGene).
Zooming in to the amino acids within the exons of the METTL3 gene, we can see the numbering is backwards, going from right to left. This can be seen in Figure 4 and is also another useful technique in determining gene and strand direction
(
 http://genome.ucsc.edu/s/education/hg38_revStrandZoom).
http://genome.ucsc.edu/s/education/hg38_revStrandZoom).
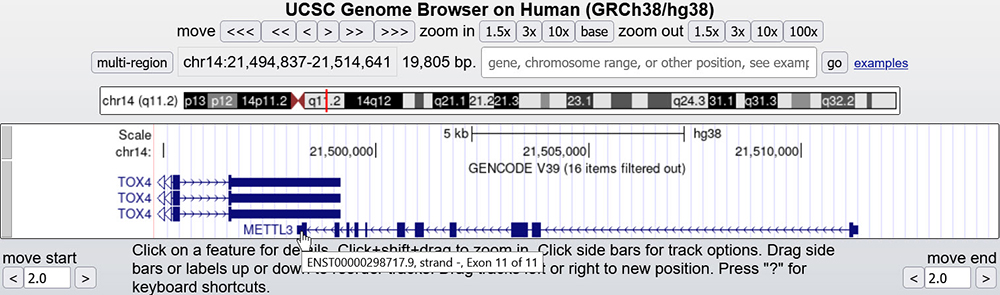
Figure 3. METTL3 gene main assembly window with exons and introns. This gene is on the bottom, or negative, strand of the DNA. We can also directly observe the arrows pointing the opposite direction.
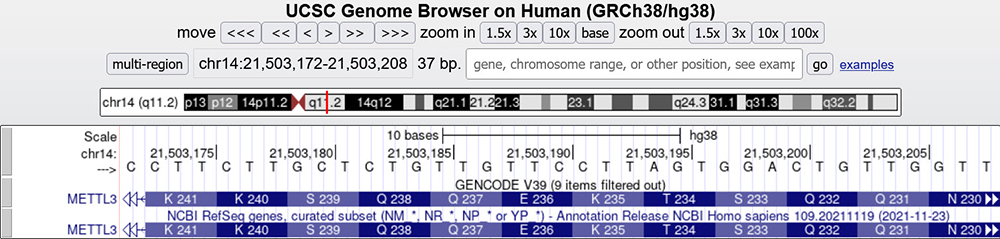
Figure 4. METTL3 gene main assembly window zoomed in on the 3rd exon. The amino acid numbering is from right to left, denoting this exon/gene is on the bottom strand. (If you are unable to see the amino acid numbering when viewing a gene at this scale, click on the gray sidebar box next to the gene, click on "Configure", check the box labeled, "Show codon numbering", and click "Apply".)
The Genome Browser makes it possible to view the DNA sequence of the bottom strand by clicking the little arrow to the left of the sequence (Figure 4), resulting in the reverse-complement of the sequence, which, when read right-to-left, represents the sequence in the same direction as transcription of the gene (Figure 5)
(
 http://genome.ucsc.edu/s/education/hg38_revStrandZoom2).
http://genome.ucsc.edu/s/education/hg38_revStrandZoom2).
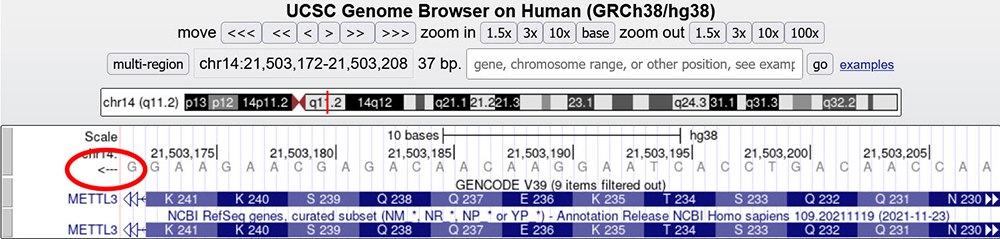
Figure 5. METTL3 gene main assembly window zoomed in on the 3rd exon. The nucleotide sequence shows the sequence of the bottom strand (red circle).
Written by Mateo Etcheveste, UCSC. Major: BS, Biomolecular Engineering