- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
WHY DON'T HUMANS HAVE TAILS?
Although they are primates, humans and other apes (hominoids) are distinct in their lack of a visible tail with only a coccyx — or tailbone — to remind us of our tailed ancient ancestors. It is very possible that as our ancestors began to acclimate to bipedal locomotion, their tail was no longer a vital morphological trait necessary to ensure balance while leaping through the thicket and springing from tree to tree. This loss of an external tail sprouted a new branch on the primate phylogenetic tree, separating a new hominoid lineage from Old World monkeys, approximately 25 million years ago. While the phenotypic difference between possessing a tail and lacking one is quite obviously observable, the genotypic contrast is not visually discernible; The Genome Browser can be used to envision the genomic change from Old World monkey to hominoid that eliminated the tail phenotype.
THE TBXT GENE
According to research performed by Xia et al., an Alu transposable element found on the hominoid TBXT gene is thought to be responsible for our loss of an external tail. There is one Alu element, "AluSx1", that is found in all primates. However, another Alu element nearby, "AluY" is only found in hominoids or primates that lack external tails. In hominoids, the two Alu elements create a loop which removes exon 6 from the transcription line. With no AluY to connect to AluSx1, primates with tails have a TBXT transcript that includes exon 6, which results in an external tail. Figure 1, which was pulled from the Xia et al. paper, illustrates the Alu loop created in hominoids and the differing transcripts between tailed and non-tailed primates.
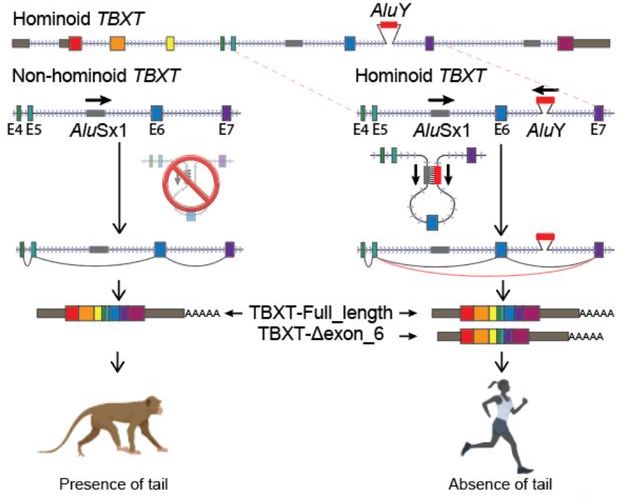
Fig 1. Pulled from the Xia et al. article (linked at the end of this paper). The hominoid TBXT gene is affected by the Alu loop created by AluSx1 and AluY, resulting in an alternately spliced transcript that lacks exon 6. Tailed primates lack the AluY element, exon 6 is transcribed, and an external tail results.
Let's use the Genome Browser as another way to visualize this genetic change. Figure 2 shows an image of the TBXT gene in its entirety. Figure 3 is the TBXT details page that can be displayed by clicking on the gene. Reading through the "Refseq Summary" reveals that TBXT plays an important role in helping to transcribe genes during mesoderm formation; The mesoderm germ layer of the embryo goes on to form the skeletal system of the fetus. The description also states that mutations within this gene are associated with chordoma, a cancer that affects the spine, and neural tube defects. This gene is obviously essential in the development of a healthy spine.
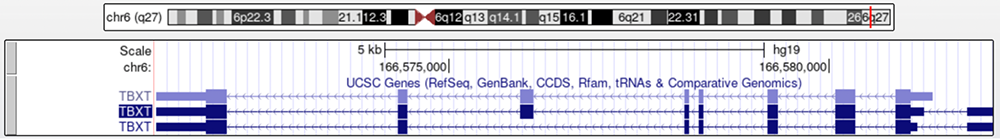
Figure 2. The human TBXT gene in its entirety. The highlighted isoform (2nd from top) is the canonical transcript.
 https://genome.ucsc.edu/s/education/hg19_TBXT
https://genome.ucsc.edu/s/education/hg19_TBXT

Figure 3. The TBXT details page that can be accessed by clicking on the TBXT gene.
 rs770199777.
rs770199777.
Turning on the "OMIM Allelic Variant Phenotypes" track confirms that mutations on this gene can have pathogenic effects. Figure 4 shows a mutation on exon 3 of TBXT that results in "sacral agenesis with vertebral anomalies," a condition where the sacral area of the spinal cord is underdeveloped, resulting in narrow skeletal and muscle morphology.
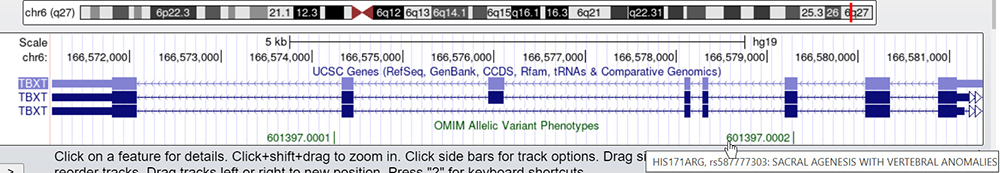
Figure 4. Mousever shows that OMIM Allele has a phenotype in the sacral region of humans,
consistent with the view that this gene is involved in formation of the lower spine.
 https://genome.ucsc.edu/s/education/hg19_TBXTomim
https://genome.ucsc.edu/s/education/hg19_TBXTomim
Turning on the RepeatMasker track reveals transposable elements on the gene, including Alu elements (which are marked within the "SINE" line of the track). Figure 5 displays the TBXT gene with the RepeatMasker track set to full. The yellow highlighted column marks the hominoid AluY element, while the turquoise highlighted column indicates the primate AluSx1 element. These two Alu elements flank exon 6, which is boxed in red. When TBXT undergoes transcription, these two Alu elements connect to one another creating a loop that stops exon 6 from being included in the mature transcript. In fact, the third transcript in the UCSC Genes track appears to be derived from transcripts that have eliminated exon 6.
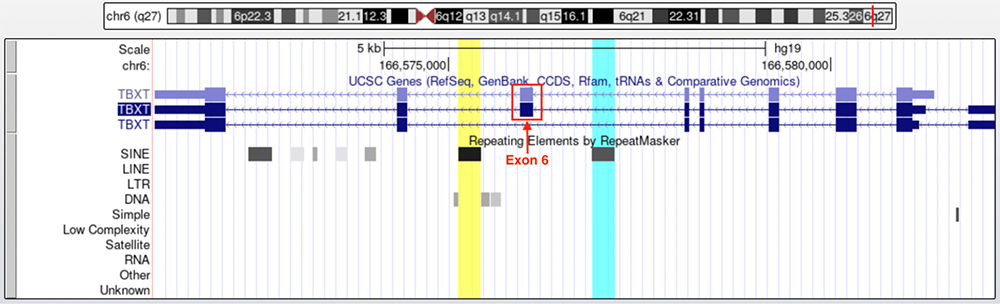
Figure 5. Yellow highlight marks AluY while turquoise highlight marks AluSx1. Exon 6 (boxed in red and labeled) is in between the two Alu elements.
 https://genome.ucsc.edu/s/education/hg19_TBXTalus
https://genome.ucsc.edu/s/education/hg19_TBXTalus
Turning on the "Multiz Alignments of 100 Vertebrates" track and selecting a variety of primate species, one can see that the AluY element, highlighted in yellow, is conserved in all non-tailed primates (Figure 6). However, in tailed primates, there is a gap in conservation where the AluY element lies. The AluSx1 element, highlighted in turquoise, is conserved in all primates.
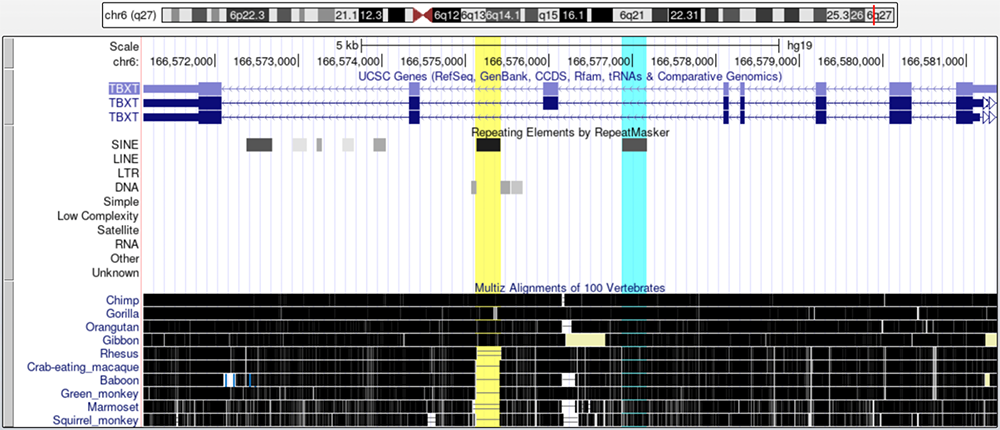
Figure 6. Conservation of the AluY (yellow) and AluSx1 (turquoise) elements in primates. AluSx1 is conserved in all primates while AluY is conserved in only non-tailed primates.
 https://genome.ucsc.edu/s/education/hg19_TBXTconservation
https://genome.ucsc.edu/s/education/hg19_TBXTconservation
Isolating the TBXT gene on the squirrel monkey genome (Figure 7) shows a lack of the AluY element but AluSx1 is present (highlighted in yellow). Figure 8 shows where the AluY element would hypothetically lie on the squirrel monkey genome if these monkeys did not have tails.
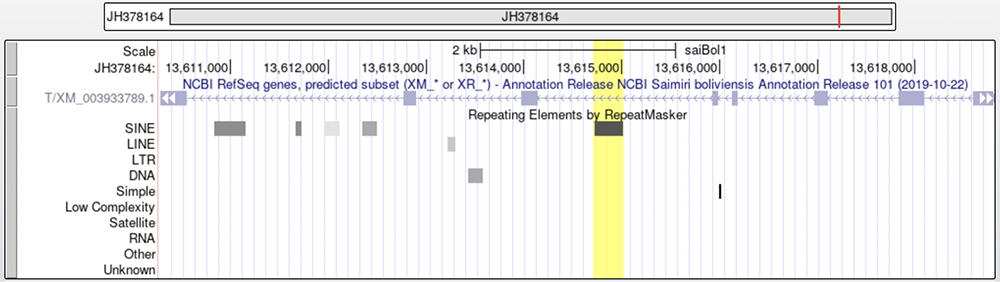
Figure 7. Squirrel monkey TBXT gene. AluSx1 is highlighted in yellow.
 https://genome.ucsc.edu/s/education/saiBol1_TBXT
https://genome.ucsc.edu/s/education/saiBol1_TBXT
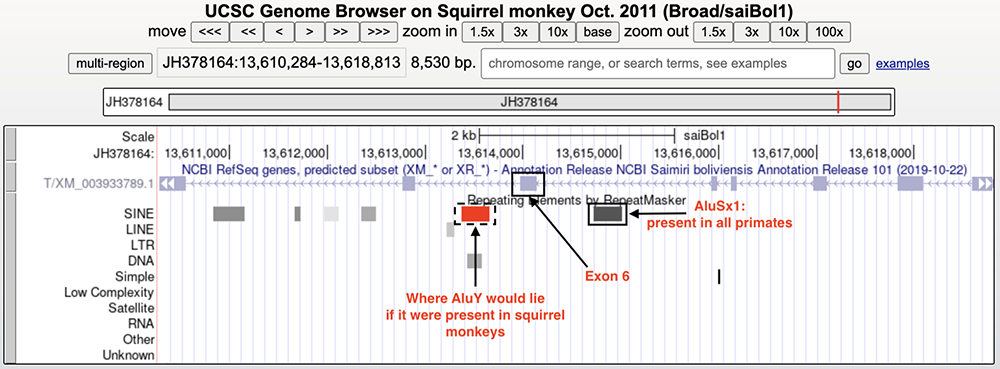
Figure 8. Squirrel monkey TBXT gene with hypothetical AluY element included.
This conversation shows us how we can learn a lot about the history and evolution of a species by looking at the genomic makeup of extant animals. We are able to pinpoint branches in a phylogenetic tree and take a deeper look into the genomic components of the species. The genomes of living animals can help us understand how we got to where we are now genetically and morphologically, especially comparing other primates seeking to learn more about humans. Our history as a species is etched into our genomes and the genomes of animals that are closely related to us.
REFERENCES
Melchor (2021). Alu leap explains why apes don't have tails
Zia, et al. (2021) (preprint) The genetic basis of tail-loss evolution in humans and apes
Written by Zoë Shmidt, UCSC. Major: BA, Biological Anthropology