- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
Track Decorators
Overview
The genome browser‘s primary way to annotate the genome uses colored rectangles ("exons" for gene tracks) linked by thin lines ("introns"), often stored as a bigBed. These were originally used for genes but then evolved to cover other types of annotations, e.g. enhancers, chromatin modifications, or single nucleotide variants. We usually call these annotations "features". Each rectangle ("exon") of a feature has the same color and individual parts cannot be highlighted. If you wanted to highlight parts of the features, traditionally this required a second track.

Track decorators allow highlighting parts of features with colors and/or symbols (glyphs/shapes) within a single track. The decorations can either be overlaid onto the feature or shown directly underneath. This is more compact than creating a separate track for these sub-features, but loses some of the abilities of normal tracks, for example, there is no right-click on the sub-features and the user cannot click these sub-features. The primary use case driving this display was protein-domains drawn on top of gene models, but we have found many other applications since then, e.g. drawing summits on ATAC peaks or highlighting selenocysteines on transcripts.
Track decorators can be shown in two styles, "block" and "glyph" style. The "block" style option can be used to color exons and introns and can display a label for them. For example, the "block" track decorator could be used to overlay protein domain boundaries on transcripts where usually one would use an entirely different track for the domains.
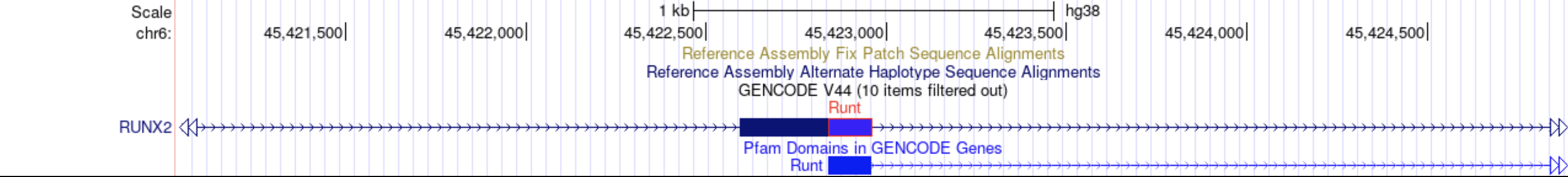
The "glyph" style option offers 8 different types of glyphs and the color of choice.

The "glyph" style option can be used to draw entirely new symbols, for example, to indicate insertion positions on the genome with small triangles. We appreciate user feedback. If you have glyph questions, glyph style requests, or have found new glyph applications, please contact our mailing list.
To use decorators in your track hub or custom tracks, you will need to create an additional bigBed file that defines the regions, colors, and glyphs.
Getting Started
Decorators can be added to BED 12+, bigBed, PSL, and bigGenePred tracks. To add decorations to a track, the decorations must first be stored in a separate bigBed file that includes extra fields to identify the decorated items. The track settings for the track must then be modified to include a pointer to that bigBed file; this can be done either in the track line for a custom track, or in the trackDb.txt file for a hub.
At present, we only support a single decorator per track, but we anticipate supporting multiple decorators in the future. For now, all decorator settings must use the prefix "decorator.default".
Hub trackDb settings
track testTrack type bigBed 12 bigDataUrl https://genome.ucsc.edu/goldenPath/help/examples/decorator/main.bb itemRgb on
The above trackDb statement provides a basic entry for a bigBed track. To add a decorator to this track, we can start by adding decorator.default.bigDataUrl as a setting, which will point to the bigBed file containing decorations.
track testTrack type bigBed 12 bigDataUrl https://genome.ucsc.edu/goldenPath/help/examples/decorator/main.bb itemRgb on decorator.default.bigDataUrl https://genome.ucsc.edu/goldenPath/help/examples/decorator/decorations.bb
Other settings are available to further configure decorators. Each may be applied to the decorator instead of the primary track by prepending "decorator.default." to the setting. For example, to set up a filterValues filter on the "keywords" field of the decorator, allowing the user to filter to any combination of three classes "Type1", "Type2", and "Type3", the following trackDb entry could be used.
track testTrack type bigBed 12 bigDataUrl https://genome.ucsc.edu/goldenPath/help/examples/decorator/main.bb itemRgb on decorator.default.bigDataUrl https://genome.ucsc.edu/goldenPath/help/examples/decorator/decorations.bb decorator.default.filterValues.keywords Type1,Type2,Type3 decorator.default.filterType.keywords multipleListOr
Please note that this would also require building an extra keywords field into the decorator
bigBed to hold those values, see Example #3 for more details. A full list of supported decorator
settings is available in the
trackDb documentation, while the next section discusses how
to build a decorator bigBed file.
Examples
Example #1: Building a decorator bigBed
A decorator bigBed file, which contains decorations for annotating another track, is very similar to our standard bigBed file format. The only difference is the addition of some extra required fields, which describe how each decoration should be drawn and what item within that other track it annotates.
The full .as format for decorator bigBed files is as follows:
string chrom; "Chromosome (or contig, scaffold, etc.)"
uint chromStart; "Start position in chromosome"
uint chromEnd; "End position in chromosome"
string name; "Name of item"
uint score; "Score from 0-1000"
char[1] strand; "+ or -"
uint thickStart; "Start of where display should be thick (start codon)"
uint thickEnd; "End of where display should be thick (stop codon)"
uint color; "Primary RGB color for the decoration"
int blockCount; "Number of blocks"
int[blockCount] blockSizes; "Comma separated list of block sizes"
int[blockCount] chromStarts; "Start positions relative to chromStart"
string decoratedItem; "Identity of the decorated item in chr:start-end:item_name format"
string style; "Draw style for the decoration (e.g. block, glyph)"
string fillColor; "Secondary color to use for filling decoration, blocks, supports RGBA"
string glyph; "The glyph to draw in glyph mode; ignored for other styles"
A copy of this file can be found here.
Valid values for the style field are "block" and "glyph". Valid glyph entries include "Circle", "Square", "Diamond", "Triangle", "InvTriangle", "Octagon", "Star", and "Pentagram". If the text isn't recognized, Circle will be used by default.
The "decoratedItem" field (chr:start-end:item_name format) captures the link between the decoration and what item in the track is being decorated. The contents of this field must be the chromosome, BED start coordinate, BED end coordinate, and item name for the decorated item (note - these are 0-based half-open BED coordinates, not 1-based fully closed coordinates. That means they are the same values as should appear in a BED file describing the decorated item).
Here is an example of how to use this format. Consider a BED item like the following in our main track:
chr1 1000 2000 MyItem 0 + 1000 2000 0 2 400,400 0,600
We can take this BED file and construct a bigBed from it as described in the bigBed documentation. After that, if we wanted to add a decoration to this item that highlighted the region from base 1200 to base 1800, we could create a corresponding item in a decorator bigBed file like this:
chr1 1200 1800 MyDecoration 0 + 1200 1800 255,0,0,255 1 600 0 chr1:1000-2000:MyItem block 255,0,0,128 Ignored
The first color, 255,0,0,255 specifies that the decoration outline will be in solid red (255 red,
0 green, 0 blue, 255 alpha). The second color 255,0,0,128, specifies that the interior of the
decoration will be shaded toward pure red, but that the decorated item will still be partially visible behind it
(due to the alpha value of 128). The chr1:1000-2000:MyItem entry describes which item in the main
track is to be annotated. In this case, it's an item with the name "MyItem" that spans chr1 from base 1000 to
base 2000. The final Ignored value is not used here because we are creating a block decoration
(a decoration that annotates a range of bases).
To add a glyph decoration to our file that marked the final base of the transcript with a green circle, we could also include the following line in our BED file of decorations:
chr1 1999 2000 MySecondDecoration 0 + 1999 2000 0,255,0,255 1 1 0 chr1:1000-2000:MyItem glyph 0,255,0,255 Circle
If we store both of those decorations in a file named input.bed, we can then build a bigBed file from it with the following command:
bedToBigBed -type=bed12+ -as=decoration.as input.bed hg38.chrom.sizes decorations.bb
Example #2: Create a Custom Track
Create a decorator bigBed custom track using the decorator bigBed file from Example #1.
-
Construct a track line that references the bigBed
and decorator bigBed file:
browser position chr1:1000-2000 track type=bigBed name="Decorators Example Two" description="bigBed with decorators" visibility=pack bigDataUrl=https://genome.ucsc.edu/goldenPath/help/examples/decorator/main.bb decorator.default.bigDataUrl=https://genome.ucsc.edu/goldenPath/help/examples/decorator/decorations.bb
- Paste the track line into the custom track page for the human assembly, hg38.
- Click the button.
Custom tracks can also be loaded via one URL line. The link below loads the same bigBed with decorators track and sets additional parameters in the URL:
https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&hgct_customText=track%20type=bigBed%20bigDataUrl=https://genome.ucsc.edu/goldenPath/help/examples/decorator/main.bb%20decorator.default.bigDataUrl=https://genome.ucsc.edu/goldenPath/help/examples/decorator/decorations.bb&position=chr1:1000-2000
Example #3: Create a decorator bigBed with extra (custom) fields
Additional fields can also be added onto the end of the .as file if you desire, though they will not be used at all by default. Adding fields allows you to set up custom filtering of which decorations are displayed. For example, to include a "keywords" field you could add the following to the end of the decoration.as file:
int numKeywords; "Number of keywords"
string[numKeywords] keywords; "Keywords for this decorator"
You would then modify your input.bed file to include an additional fields at the end of each line, detailing which keywords apply to each of the decorations.
chr1 1200 1800 MyDecoration 0 + 1200 1800 255,0,0,255 1 600 0 chr1:1000-2000:MyItem block 255,0,0,128 Ignored 1 Type1 chr1 1999 2000 MySecondDecoration 0 + 1999 2000 0,255,0,255 1 0 1 chr1:1000-2000:MyItem glyph 0,255,0,255 Circle 2 Type2,Type3
After rebuilding the decorations.bb file, you could then use the filterValues version of the trackDb entry from the Hub trackDb settings section to create your track.
Troubleshooting
If you get an error when you run the bedToBigBed program, please check your input BED file for
data coordinates that extend past the end of the chromosome. If these are present, run the
bedClip program
(available here) to remove the problematic
row(s) in your input BED file before using the bedToBigBed program.