Description
This track shows data from
Single cell RNA sequencing of human liver reveals distinct intrahepatic
macrophage populations. Liver tissue was analyzed using droplet-based
single-cell RNA-sequencing (scRNA-seq) and subsequent clustering distinguished 20
hepatic cell populations based on their identified marker genes found in
MacParland et al., 2018.
There are three bar chart tracks in this track collection with liver cells
grouped by either broad cell type
(Liver Broad), specific cell type
(Liver Cells) and donor
(Liver Donor). The default track displayed is
Liver Cells.
Display Conventions
The cell types are colored by which class they belong to according to the following table.
| Color |
Cell classification |
| immune |
| endothelial |
| fibroblast |
| epithelial |
| stem cell |
| hepatocyte |
Cells that fall into multiple classes will be colored by blending the colors associated
with those classes. The colors will be purest in the
Liver Cells subtrack,
where the bars represent relatively pure cell types. They can give an overview
of the cell composition within other categories in other subtracks as well.
Relevant Figures From MacParland et al., 2018
Contribution of cells from each liver sample to each cell cluster. Note that
the liver number corresponds to the donor number (e.g. Liver 1 = Donor 1).
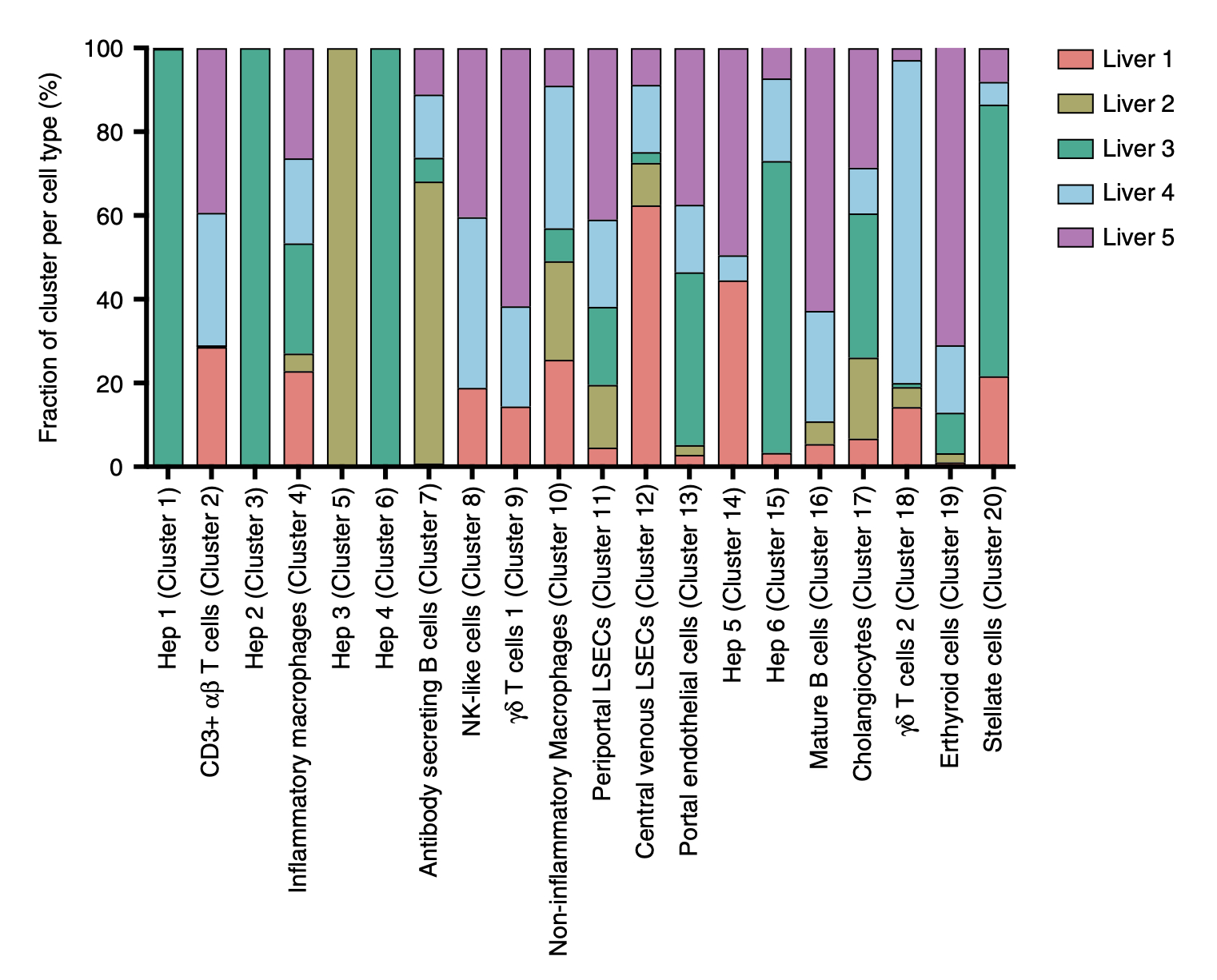 MacParland et al. Nat
Commun. 2018. / CC BY 4.0
MacParland et al. Nat
Commun. 2018. / CC BY 4.0
t-SNE plot of human liver resident cells colored by source donor (Liver 1-5)
and labeled with cluster number.
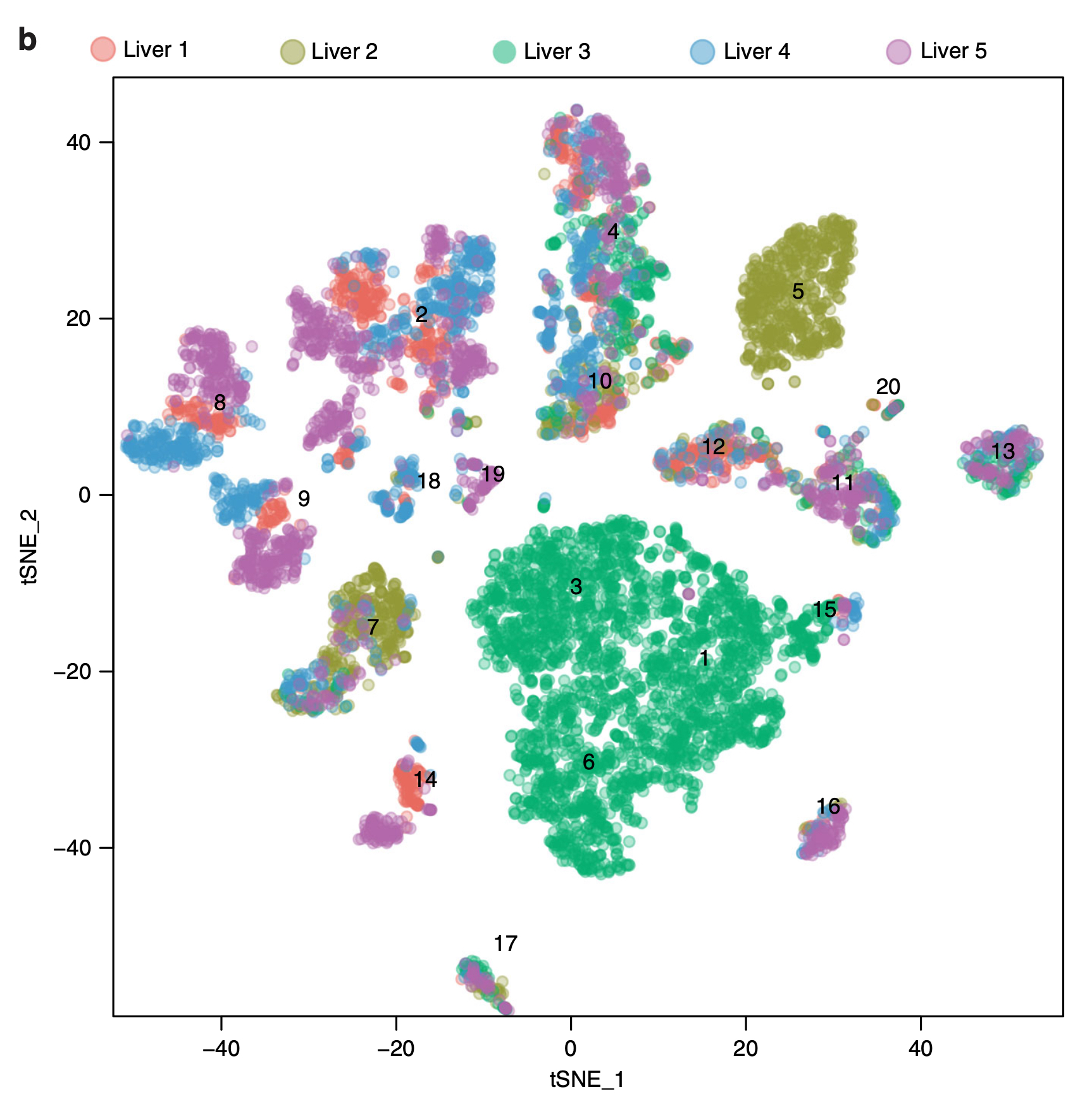 MacParland et al. Nat
Commun. 2018. / CC BY 4.0
MacParland et al. Nat
Commun. 2018. / CC BY 4.0
Method
Fresh liver samples were taken from 5 neurologically deceased donors (NDD)
deemed acceptable for liver transplantation. The caudate lobe of the liver was
surgically separated and flushed with HTK solution to leave only tissue
resident cells that were used to prepare a cell suspension for scRNA-seq
analysis. Samples were prepared using 10x Genomics 3' v2 library kit and
sequenced on the Illumina HiSeq 2500. A total of 8,444 transcriptional profiles
were obtained for organ specific and non-organ specific cells from healthy
hepatic tissue.
The cell/gene matrix and cell-level metadata was downloaded from the
UCSC Cell Browser.
The UCSC command line utility matrixClusterColumns, matrixToBarChart, and bedToBigBed were used
to transform these into a bar chart format bigBed file that can be visualized. The coloring
was done by defining colors for the broad level cell classes and then using another UCSC utility,
hcaColorCells, to interpolate the colors across all cell types. The UCSC utilities can be found on
our download server.
Data Access
The raw bar chart data can be
explored interactively with the Table
Browser or the Data Integrator. For
automated analysis, the data may be queried from our REST API. Please refer to our mailing
list archives for questions, or our Data Access FAQ for more
information.
Credit
Thanks to Sonya MacParland and to the many authors who worked on producing and
publishing this data set. The data were integrated into the UCSC Genome Browser
by Jim Kent and Brittney Wick then reviewed by Daniel Schmelter. The UCSC work
was paid for by the Chan Zuckerberg Initiative.
References
MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares
I et al.
Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations.
Nat Commun. 2018 Oct 22;9(1):4383.
PMID: 30348985; PMC: PMC6197289
|